Master’s Project: Decoding Thoughts
Decoding Mental Representation of of Nouns and Verbs
1. Introduction
When understand or produce sentences there is a mental representation of the content of that sentences. Question is: where is that representation?
In this project we try to “decode” this representation by using Machine Learning algorithms. The idea is, if we can decode them with above average accuracy, then we know something about the representation is present at that location and time.
There are two candidates for where and when such representations may become available. One is based on a famous model by Friederici (2011) which predicts the involvement of temporal lobe at around 600 ms after the stimulus was presented (see below). Another is a more recent prediction made by Pylkkänen (2020). This model predicts earlier activation of frontal region, an area associated with episodic memory.
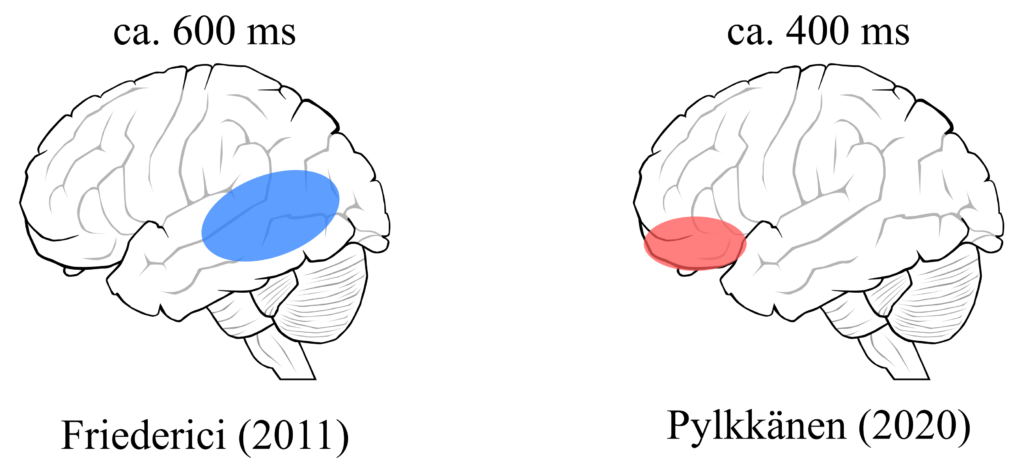
These two predictions are very different, therefore we have a good chance of spotting the difference.
1.1. Magnetoencephalography: What is it? Why use it?
Magnetoencephalography or MEG is a form of neuroimaging technique. It measures minuscule magnetic field created by electric currents inside our head.
There are a few advantages of using MEG over other neuroimaging techniques.
MEG is fast: unlike fMRI, which takes hundreds of milliseconds to take snapshots of the neural activity, MEG can measure activity in realtime.
MEG has better localisation: compared its cousin electroencephalography (EEG), MEG can localise the source more accurately.
Because we want to know when and where of neural activities MEG is the perfect tool to use for this project.
1.2. Multivariate Pattern Analysis (a.k.a. Machine Learning)
In order to analyse the data we have used multivariate pattern analysis (MVPA). This may be more familiar to you as Machine Learning.
1.2.1. Why Use Machine Learning?
Human brain has over 1011 neurons. Each neuron has thousands of synapses that connect to other neurons. Trying to understand every tiny detail is beyond human capability. The traditional solution to this problem is to average out the data to see “peaks” of activations. However, this means throwing away a large portion of data.
Machine Learning can help us solve this problem without throwing away information by detecting patterns automatically without human intervention. We only need to care about global patterns.
1.2.2. Data Analysis Pipeline
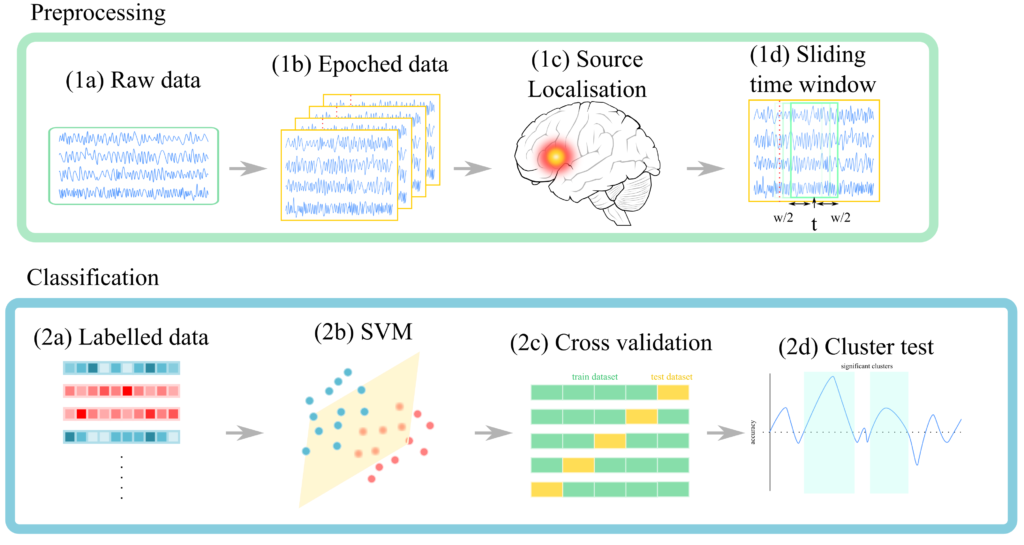
In this project we use following processing pipeline:
(1a) Raw data: data directly obtained from the experiment
(1b) Epoched data: “epoching” is simply the act of cutting up continuous data by time-locking to some event (e.g. stimulus presentation). Thus, we will have as many data points as there are events.
(1c) Source Localisation: this simply means estimating the location of the activation source from measured activities. I defer explanation to MNE-python tutorials.
(1d) Sliding time window: we want to know at which time point information was available. In order to do this we take data from a small (sliding) window.
(2a) Labelled data: each event has corresponding stimulus (e.g. noun, verb etc.) We can pair individual data point (now flattened) with these stimuli identity (i.e. labels) to perform supervised learning.
(2b) Classification: we have used linear support vector machine (linear SVM). The reason is because our dataset is very small (240 data points). Linear SVM tend to do well with small datasets like this one. It is also relatively easy to interpret compared to Deep Learning algorithms.
(2c) Cross validation: k-fold cross validation is performed
(2d) Cluster test: in order to assess whether the decoding performance is above average or not, we performed non-parametric cluster test. This will give us the p-values to threshold the results.
Our pipeline is summarised below.
You can now continue to read about pilot study analysis or MOUS dataset analysis below.
A. Final Thesis
The final thesis submitted can be downloaded from here.
B. Data
Please note that due to data protection laws I cannot share the raw pilot data used for the analyses.
Mother of All Unification dataset can be accessed after registration process here.
C. Source Code
Scripts used in this project are available here.
D. References
Friederici, A. D. (2011). The brain basis of language processing: from structure to function. Physiological reviews, 91(4), 1357-1392.
Pylkkänen, L. (2020). Neural basis of basic composition: what we have learned from the red–boat studies and their extensions. Philosophical Transactions of the Royal Society B, 375(1791), 20190299.
Schoffelen, J. M., Oostenveld, R., Lam, N. H., Uddén, J., Hultén, A., & Hagoort, P. (2019). A 204-subject multimodal neuroimaging dataset to study language processing. Scientific data, 6(1), 1-13.
